Metagenomes are already complex mixtures of species that can be hard to describe. But many modern studies actually have large collections of metagenomes, for example those looking at the human gut or skin microbiomes in relation to diseases. These datasets have huge potential, but pose a challenge for visual analytics. To make the problem more tractable, we adapt learning methods from Natural Language Processing. Making the analogy of each metagenome as a document, and the species within the metagenomes as words in the documents, we can apply Topic Models, which find common, interpretable themes among large collections of text. In our context, this can show us what communities of microbes tend to appear together.
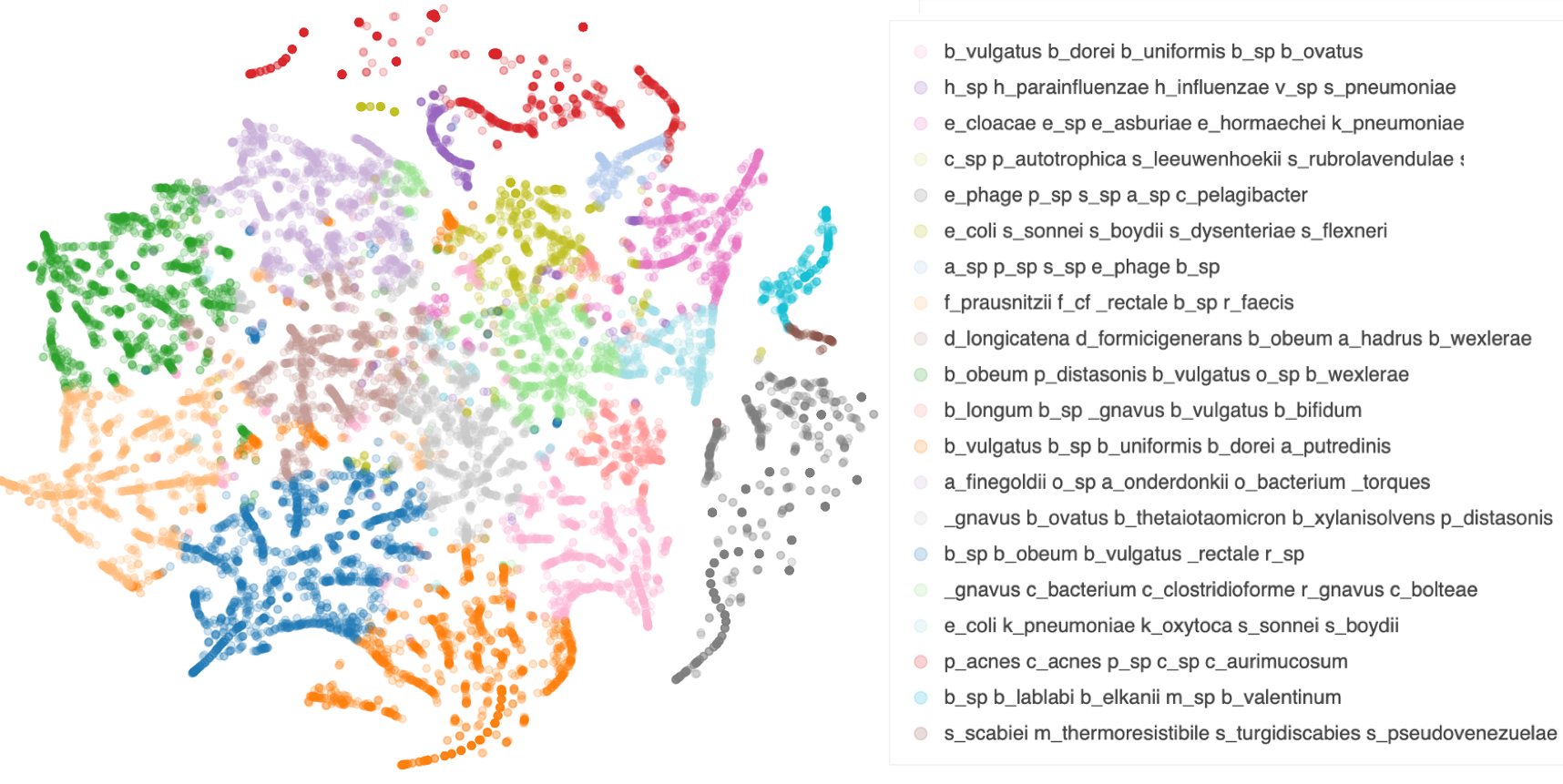
This provides a label of which microbial communities dominate each metagenomic sample (the coloring of the points above), and provides lists of which microbes make up each of those communities. Combined with metadata, this could be a powerful tool for exploring these massive datasets and for gaining insights about what drives differences in microbiomes and other metagenomes.